| Photoiupac home page | Discussion | Photobiology.com home |
DNA in dormant Bacillus subtilis spores exposed to solar radiation accumulates strand breaks and cyclobutane pyrimidine dimers in addition to spore photoproduct
Tony A. Slieman and Wayne L. Nicholson*
Department of Veterinary Science and Microbiology, University of Arizona, Tucson, AZ 85721
*Corresponding author. Telephone (520) 621-2157, Fax (520) 621-6366, electronic mail address WLN@u.arizona.edu
SUMMARY
While UV irradiation of dormant B. subtilis endospores results mainly in formation of the "spore photoproduct" (SP) 5-thyminyl-5,6-dihydrothymine, genetic evidence indicates that additional DNA photoproduct(s) may be formed in spores exposed to solar UV-B and UV-A radiation (Xue,Y. and W.L. Nicholson, Appl. Environ. Microbiol. 62: 2221-2227. 1996). The presence of double-strand (ds) breaks, single-strand (ss) breaks, cyclobutane pyrimidine dimers (Py<>Py) and apurinic/apyrimidinic (AP) sites in spore DNA was determined under solar UV irradiation conditions using a combination of enzymatic probes and neutral- or alkaline-agarose gel electrophoresis. DNA from spores exposed to full-spectrum sunlight (UV-B + UV-A) accumulated single-strand (ss) breaks, double-strand (ds) breaks and cyclobutane pyrimidine dimers (Py<>Py), whereas spores exposed to sunlight from which the UV-B component had been filtered ("UV-A sunlight") accumulated ss breaks and ds breaks only. Heating of the spores during solar exposures did not noticeably damage spore DNA. AP sites were not detected in spore DNA under any exposure condition used.
INTRODUCTION
The bacterial endospore is a highly-evolved structure capable of maintaining the bacterial genome in a protected, viable state for extended periods (reviewed in Setlow, 1995). Spores of the common soil bacterium Bacillus subtilis have proven to be a particularly fruitful system for field studies of the consequences of long-term cellular exposure to solar radiation, due to:
The nucleotide excision repair (NER) and spore photoproduct (SP) lyase DNA repair pathways are major determinants of spore resistance to solar radiation, as mutant B. subtilis spores lacking both repair systems exhibit extreme sensitivity not only to laboratory UV-C radiation (Munakata, 1969), but also to the UV wavelengths present in sunlight (Munakata 1981; 1989; 1993; Quintern et al., 1994; Tyrrell, 1978; Wang, 1991; Xue and Nicholson, 1996).
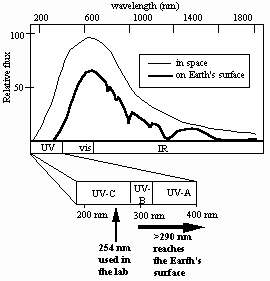
How well does the current laboratory model describe spore UV resistance in the environment? Solar radiation reaching the Earth’s surface is considerably more complex than artificially-produced monochromatic 254-nm UV,consisting of a mixture of UV, visible, and infrared radiation, the UV portion spanning approximately 290-400 nm (the so-called UV-B and UV-A portions of the UV spectrum) (Urbach and Gange, 1986) (Fig. 1).

Fig. 1. Solar spectrum in space (thin line) and at Earth’s surface (thick line).
In agreement with the current laboratory model, it has been well- documented that DNA in spores exposed either directly to solar radiation, or in the laboratory to UV wavelengths present in sunlight, accumulate mainly "spore photoproduct" (SP; the thymine dimer 5-thyminyl-5,6-dihydrothymine) as the major UV photoproduct (Tyrrell, 1978) (Fig. 2).
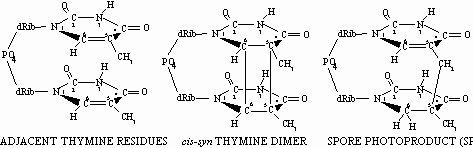
Fig. 2. Structures of adjacent thymines in DNA (left), cyclobutane thymine dimer (center) and spore photoproduct (rught); dRib=2’-deoxyribose.
In contrast to the laboratory model, however, in spores exposed to UV-C, UV-B, or full-spectrum sunlight, a shift towards NER is observed in the relative contributions of NER and SP lyase to spore UV resistance (Xue and Nicholson, 1996). These results were interpreted to indicate that environmentally-relevant UV wavelengths also induced non-SP photoproduct(s) in spore DNA which were preferentially repaired by NER (Xue and Nicholson, 1996). In addition, exposure of spores of NER- or SP lyase-deficient mutant B. subtilis strains to UV-A sunlight consisting of wavelengths >320 nm resulted in lethal damage which was in large part repaired neither by NER nor by SP lyase (Xue and Nicholson, 1996). Collectively, the data indicated that exposure of spores to solar radiation may have produced DNA photoproduct(s) in addition to SP.
What might be the nature of these putative photoproducts? As suggested previously (based upon considerations discussed extensively in Xue and Nicholson, 1996), cyclobutane-type pyrimidine dimers (Py<>Py), AP sites and strand breaks in DNA are possible candidates for solar radiation-induced damage in spore DNA. The present communication describes experiments designed to test this hypothesis.
MATERIALS AND METHODS
Bacterial strains and culture conditions. The B. subtilis strain used throughout this study was WN356 [metC14 sul thyA1 thyB1 trpC2 uvrB42 D(splAB)::ermC] which lacks NER and SP lyase activities and has been described previously (Nicholson et al., 1997). Spores were routinely prepared by growth and sporulation for 48-72 hr. at 37˚C in nutrient broth sporulation medium (NSM)(Schaeffer et al., 1965). Suspensions of sporulated cultures were treated with lysozyme (10mg/ml) to remove vegetative cells and further purified by repeated washing in buffers and centrifugation, followed by heat shock (80˚C for 10 min) as described in detail previously (Nicholson and Setlow, 1990). The resulting spores were ascertained by phase-contrast microscopy to be > 99.9% pure.
Solar exposure. Suspensions of purified spores (5.2 x 1010 colony forming units [cfu] total) were layered onto the bottom halves of sterile 10-cm diameter polystyrene Petri dishes and air-dried at 55˚C. The resulting dried spore samples were subjected to sunlight during the daily period of maximal solar intensity; local noon was calculated for the longitude of Tucson, AZ (111˚ 2’ W) using the Voyager II computer program (Carina Software, San Leandro, CA). For exposure to full-spectrum sunlight, samples were covered with a single layer of Saran wrap, which transmits essentially all solar UV wavelengths (Xue and Nicholson, 1996). Exposure of spores to sunlight from which the UV-B portion of the spectrum had been removed (called here "UV-A sunlight") was accomplished by covering the samples with a 1.25-cm (1/2 inch-) thick glass plate, previously determined to completely block transmission of UV wavelengths shorter than 325 nm (Xue and Nicholson, 1996). Solar dosimetry was performed using a UVX radiometer fitted with the appropriate UV-B and UV-A probes and readings were taken under the same shielding materials which covered the spore samples. Dose rate readings (reported in J / m2 . sec) were taken at hourly intervals and the average of two successive readings was used to estimate the total UV dose (in J / m2) received by samples during the interval. In order to obtain the desired solar UV dose (especially for samples exposed to UV-A sunlight) it was often necessary to carry out exposures over the course of several days.
During the course of solar exposures, ambient temperatures exceeding 70˚C were routinely recorded (Fig. 3).
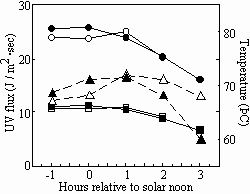
Fig. 3. Solar dosimetry performed on 21 July, 1999 (open symbols) and 22 July, 1999 (solid symbols). Temperature (triangles), UV-A flux (circles) and UV-B flux (squares) were recorded.
In order to control for potential DNA damage caused by heat, spore samples shielded with a single layer of aluminum foil were exposed to solar radiation in parallel and treated identically to account for heat damage. At the end of each exposure period, samples were transported to the laboratory and stored at room temperature in the dark until the following exposure period. Spore survival to solar radiation or heat was performed essentially as described previously (Xue and Nicholson, 1996).
DNA isolation, manipulation, and electrophoresis. Exposed dried spore samples were resuspended in 10 ml phosphate buffered saline (PBS; 10 mM potassium phosphate, 150 mM NaCl, pH=7.4) and spores were harvested from Petri dishes using a sterile spatula. The resulting spore suspensions were collected by centrifugation, resuspended in decoating solution (8M urea, 15mM Tris base, 1% SDS, 50mM DTT) and incubated at 60˚C for 90 min to remove the protein coat. Decoated spores were washed, centrifuged, and resuspended three times with STE buffer (10 mM Tris-HCl, pH 8, 10 mM EDTA, 150 mM NaCl) and once with lysis solution (50 mM NaCl, 100 mM EDTA). Spores were then lysed and chromosomal DNA was extracted and purified as previously described (Cutting and Vander Horn, 1990). To detect Py<>Py, DNA was first digested with phage T4 endonuclease V (Endo V)(Epicentre Technologies, Madison, WI), which cleaves the phosphodiester backbone 5’ to Py<>Py. To detect apurinic/apyrimidinic (AP) sites, DNA was first digested with Endonuclease IV (Endo IV) (Epicentre Technologies) (Ljungquist, 1976). Neutral agarose gel electrophoresis of DNA was performed by standard techniques (Sambrook et al., 1989). In order to detect ss breaks generated in DNA either directly by UV treatment or as a result of Endo V or Endo IV cleavage at Py<>Py or AP sites, DNA was denatured with 300 mN NaOH (final concentration) and electrophoresed at 4˚C through 0.8% alkaline agarose gels with buffer recirculation essentially as described previously (Sambrook et.al., 1989). Migration of DNA was determined relative to a set of molecular weight standards ranging in size from 0.5 - 12 kb (1 Kb Ladder, Life Technologies, Gaithersburg, MD).
RESULTS
In a control experiment, DNA extracted from unirradiated spores of strain WN356 and separated on either 0.8% neutral- or alkaline-agarose gels beside high-molecular-weight markers (phage l digested with HindIII) demonstrated either 23-kbp ds fragments or 23-kb ss fragments, respectively (data not shown); thus, DNA extracted and purified from spores was uniformly sheared to approx. 23-kbp ds fragments, and suffered no detectable additional ss breaks during the purification procedure.
Spores of strain WN356 were exposed to full-spectrum solar radiation, UV-A solar radiation, or solar heating. During the exposure period, measurements of temperature, and UV-B and UV-A flux were recorded at hourly intervals. Chromosomal DNA isolated from exposed spores was then probed for ds breaks by neutral agarose gel electrophoresis and ss breaks by denaturation in alkali followed by alkaline-agarose electrophoresis. Pretreatment of DNA with T4 Endonuclease V or Endonuclease IV was used to detect presence of Py<>Py or AP sites, respectively. The results are shown in Fig. 4.
Spores of strain WN356 covered with Saran wrap were exposed to full-spectum solar radiation on the 4th and 5th of August, 1998. The total dosage for the full spectrum irradiated spores was determined to be 8.23 x 105 J/m2 UV-A and 3.53 x 105 J/m2 UV-B, respectively. Exposure of spores to full spectrum sunlight resulted in the formation of ss breaks and Py<>Py in chromosomal DNA of spores (Fig. 4B). Ds breaks were also detected (Fig. 4A), but no AP sites were present (data not shown). Despite the fact that the temperature exceeded 70˚C during the experiments, DNA damage in spores exposed to full-spectrum sunlight or UV-A sunlight was not caused by heat, as spores exposed in parallel to the same heat regimen but shielded from solar radiation exhibited no detectable DNA damage (Figs. 4A and 4B).
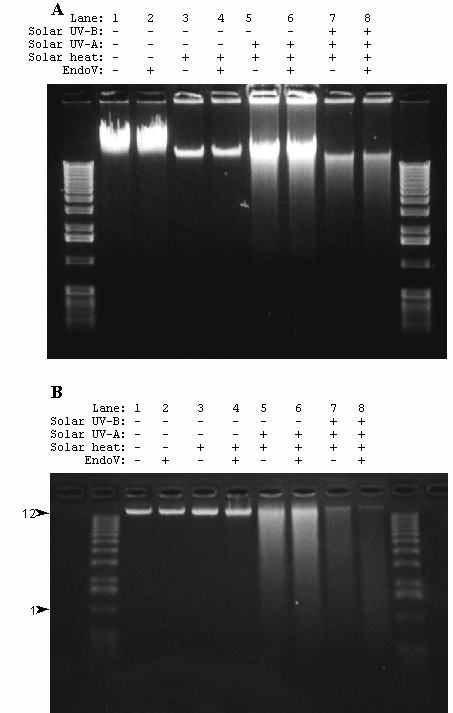
Fig. 4. Chromosomal DNA extracted from spores of strain WN356 exposed to UV-A sunlight on 5-16 October 1998 and full spectrum sunlight on 4-5 of August 1998 was electrophoresed through a neutral 0.8% agarose gel (A) or a 0.8% alkaline-agarose gel (B). Spores are: not exposed (lanes 1 and 2); exposed in parallel to heat only (lanes 3 and 4); exposed in parallel to UV-A sunlight only (1.1 x 10 6 J / m2, lanes 5 and 6); exposed to full spectrum sunlight (8.23 x 105 J/m2 UV-A + 3.53 x 105 J/m2 UV-B) (lanes 7 and 8). Isolated DNA was treated with Endo V before electrophoresis (lanes 2, 4, 6, and 8). M, molecular weight markers. The 12 kb and 1 kb markers are indicated with arrowheads..
On clear days from 5 to 16 October 1998, spores of strain WN356 were exposed under 1.25-cm plate glass to UV-A sunlight to a total dosage of 1.1 x 106 J/m2. DNA extracted from spores exposed to UV-A sunlight and electrophoresed through 0.8% neutral and 0.8% alkaline-agarose gels was observed to contain ds breaks (Fig. 4A) and ss breaks (Fig. 4B), but virtually no Py<>Py (Fig. 4B) or AP sites (data not shown). Again, damage to spore DNA was due to direct exposure to solar radiation and not to heat, as a parallel set of spores shielded from UV by aluminum foil but exposed to the same temperature regimen accumulated no detectable DNA damage (Fig. 4).
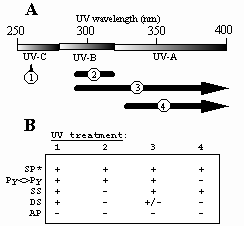
Fig.5. A. UV treatments to which spores have been exposed. 1, artificial UV-C (254 nm); 2, artificial UV-B (290-310 nm); 3, full-spectrum sunlight (>290 nm); 4, "UV-A" sunlight (>325 nm). B. Summary of B. subtilis spore photochemistry at different artificial and environmental UV wavelengths. UV treatments are numbered as in part (A). SP, spore photoproduct; Py<>Py, cyclobutyl pyrimidine dimers; SS, single-strand breaks; DS, double-strand breaks; AP, apurinic/apyrimidinic sites. +, damage detected; +/-, damage barely detectable; -, damage not detected. *SP data are from Tyrrell (1978).
CONCLUSIONS
In order to understand the photochemistry of spore DNA in the environment as compared to the artificial laboratory model, spores were irradiated with full-spectrum sunlight and UV-A sunlight. In addition to the well-characterized spore photoproduct (SP), spores were observed to accumulate ds breaks, ss breaks, and Py<>Py (Fig. 5).
This study clearly showed that B. subtilis spores accumulated Py<>Py in addition to the predominant SP when subjected to environmentally-relevant full-spectrum solar radiation. Furthermore, both full-spectrum solar UV and UV-A alone induced ds breaks and ss breaks. The results suggest that it is the UV-B component of sunlight which is responsible for Py<>Py formation in spore DNA. In contrast, no apurinic/apyrimidinic sites were detected in any of our DNA preparations as revealed by digestion with Endo IV (data not shown).
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health (GM47461) and the Arizona Agricultural Experimental Station (USDA-HATCH-ARZT-136753-H-02-116) to W.L.N.
REFERENCES
*Cutting, SM, and P.B. Vander Horn. 1990. Genetic analysis, p. 27-74. In C.R. Harwood and S.M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley and Sons, Sussex, England.
* Ljungquist, Siv. 1976. A new endonuclease from Escherichia coli acting at apurinic sites in DNA. J. Biol. Chem. 262: 2808-2814.
*Munakata, N. 1969. Genetic analysis of a mutant of Bacillus subtilis producing ultraviolet-sensitive spores. Mol. Gen. Genet. 104: 258-263.
*Munakata, N. 1981. Killing and mutagenic action of sunlight upon Bacillus subtilis spores: a dosimetric system. Mutat. Res. 82: 263-268.
*Munakata, N. 1989. Genotoxic action of sunlight upon Bacillus subtilis spores: monitoring studies at Tokyo, Japan. J. Radiat. Res. 30: 338-351.
*Munakata, N. 1993. Biologically effective dose of solar ultraviolet radiation estimated by spore dosimetry in Tokyo since 1980. Photochem. Photobiol. 58: 386-392.
*Nicholson, W.L., L. Chooback, and P. Fajardo-Cavazos. 1997. Analysis of spore photoproduct lyase operon (splAB) structure and function using targeted deletion-insertion mutations spanning the Bacillus subtilis ptsHI and splAB operons. Molec. Gen. Genet. 255: 587-594.
*Nicholson, W.L. and P. Fajardo-Cavazos. 1997. DNA repair and the ultraviolet radiation resistance of bacterial spores: from the laboratory to the environment. p. 125-140. In, S. Pandalai (ed.) Recent Research Developments in Microbiology, vol. 1. Research Signpost, Trivandrum, India.
*Nicholson, W.L. and P. Setlow. 1990. Sporulation, germination, and outgrowth, pp. 391-450. In, Harwood, C.R. and Cutting, S.M. (eds). Molecular biological methods for Bacillus. John Wiley and Sons, Sussex, England.
*Quintern, L.E., M. Puskeppeleit, P. Rainer, S. Weber, S. el Nagger, U. Eschweiler, and G. Horneck. 1994. Continuous dosimetry of the biologically harmful UV radiation in Antarctica with the biofilm technique. J. Photochem. Photobiol. B: Biol. 22:59-66.
*Sambrook, J., E.F. Fritsch, and T. Maniatis (ed.). 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, NY, U.S.A.
*Schaeffer, P., J. Millet, and J.-P. Aubert. 1965. Catabolic repression of bacterial sporulation. Proc. Natl. Acad. Sci. USA 54: 704-711.
*Setlow, P. 1995. Mechanisms for the prevention of damage to DNA in spores of Bacillus species. Annu. Rev. Microbiol. 49: 29-54.
*Tyrrell, R.M. 1978. Solar dosimetry with repair deficient bacterial spores: action spectra, photoproduct measurements and a comparison with other biological systems. Photochem. Photobiol. 27: 571-579.
*Urbach, F. and R.W. Gange (ed). 1986. The biological effects of ultraviolet A radiation. Praeger Publishers, New York.
*Wang, T.-C.V. 1991. A simple convenient biological dosimeter for monitoring solar UV-B radiation. Biochem. Biophys. Res. Commun. 177:48-53.
*Xue, Y. and W.L. Nicholson. 1996. The two major spore DNA repair pathways, nucleotide excision repair and spore photoproduct lyase, are sufficient for the resistance of Bacillus subtilis spores to artificial UV-C and UV-B but not to solar radiation. Appl. Environ. Microbiol. 62: 2221-2227.