XVIII IUPAC Symposium on Photochemistry, July
22-27, 2000
Virtual Photochemistry Poster
Session
Molecular Pathways of Electron Transfer.
Tunnelling Effect and
Van der Waals Contacts in the Cytochrome
f-Plastocyanin Complex
M. Fragata
Université du Québec à Trois-Rivières,
Département de chimie-biologie, Section de chimie,
Trois-Rivières, Que, G9A 5H7, Canada
fragata@uqtr.uquebec.ca
NB: As
I will travel out of Trois-Rivières from July 17 to August 3,
to places where most likely the Web shall not
be available, I would be much obliged to
those of you who want to comment on this work
to write your questions, nevertheless!.
At my return from the 'web wilderness',
I will answer to everybody with great pleasure.
TABLE OF CONTENTS
Abstract
1. Introduction
2. Methods
3. Tunnelling
effect vs. Van der Walls contacts
in the
cytochrome f-plastocyanin complex
( i) Structure-function
model I
(ii) Structure-function
model II
4. Concluding remarks
References
Acknowledgements
Abstract
back
A study was undertaken of the molecular pathways of electron transfer
in the cytochrome f-plastocyanin (cyt f-Pc )
complex. Between the Fe-atom in cyt f and the the Cu-atom in Pc,
the calculated centre-to-centre distances of electron transfer distances
(De) are 7.2 Å from the Fe-atom to tyrosine 1 (Y1) in cyt f, and
De = 5.9 Å from Y1 to the copper atom (Pc). Within Pc, De =
7.0 Å between cysteine 84 (C84) and
serine 85 (S85) and 5.2 Å between S85(OH) and tyrosine 83 [Y83(OH)].
A calculated alternative route of electron transfer in Pc is between C84(SH)
and the pi-electron system in Y83 where De is ~7.3 Å.
On the other hand, the electron transfer distance between the Cu-atom and
C84(SH) is approximately 2.6 Å, i.e., the coordination distance from
the copper atom to the SH group in C84. De = 2.6 Å is
clearly quite small in relation to the 5 to ~7 Å determined for the
various electron transfer pathways in cyt f-Pc. In this respect,
it is noted first that electron transfer distances higher than about 5
Å are in general a good indication of electron transfer occurring
by tunnelling efffect, that is, by a quantum jump from one electronic state
to another. On the contrary, it is reasonable to assume that electron
transfer reactions at distances as small as 2.6 Å might simply take
place via Van der Waals contacts. This is a convincing argument to
suggest the coexistence of different molecular mechanisms of electron transfer
in the cyt f-Pc complex. Such coexistence of molecular states
has far reaching consequences so far as it presupposes the function in
cyt f-Pc of adiabatic and non-adiabatic electron transfer processes which
have different temperature dependence, or requirements.
1. Introduction
back
Electron transfer between redox proteins is of fundamental
importance in living cells and organisms such as the chloroplasts of green
plants and algae. This question presents also a practical interest
in the construction of efficient batteries for solar energy conversion
into chemical or electrical energy. Nevertheless, these devices are
often plagued with difficulties inherent to energy losses due to back reactions.
In natural systems, however, this question has apparently been solved with
a complex, yet elegant system of molecular pathways leading to a remarkably
efficient transfer of electrons through molecular distances that are often
very long, e.g., ~ 20-25 Å or more (see data discussed in [1]).
In this perspective, the electron transfer mechanisms in the photosynthetic
membrane of the chloroplast made the object of a large number of works
(see, e.g., [2-4]). In spite of this, the molecular pathways
of the photosynthetic electron transfer are not yet clearly understood.
In this study, we discuss
first the most relevant pathways of electron
transfer in the cytochrome f-plastocyanin complex. Secondly,
we shall try to characterize the molecular pathways of electron transfer
by tunnelling effect and via Van der Walls contacts that may be at the
origin, and coexistence, of non-adiabatic and adiabatic electron
transfer in cyt f-Pc.
2. Methods
back
The molecules used in this work were obtained from the
Protein Data Bank [5], that is, 5pcy.pdb [plastocyanin (Pc)]
and 1ctm.pdb [cytochrome f (cyt f)]. The cyt f-Pc complex
was modelled [6] using the computer graphics program TURBO-FRODO
[7,8], and the structures were further refined with the X-PLOR program
(version 3.1) for the energy minimization (see details in [9]).
The atomic distances and other molecular details in the in Pc and the cyt
f-Pc complex were determined with the WebLab ViewerPro software from Molecular
Simulations Inc. (San Diego, CA). Other calculations were performed
with Maple (version V) from Waterloo Maple Inc. (Waterloo, ON), and Origin
(version 5) from Microcal Software, Inc. (Northampton, MA).
3. Tunnelling
effect vs. Van der Walls contacts
in the cytochrome f-plastocyanin complex
back
(i) Structure-function model I
In higher plants chloroplasts the cytochrome b6f (cyt b6f)
complex transfers electrons between the photosystem II complex and the
P700 reaction center in the photosystem I (PSI) complex. From cytochrome
f (cyt f) in cyt b6f to PSI, the electrons are shuttled by the mobile redox
carrier plastocyanin (Pc) [10-13]. Fig. 1 shows a detail
of the electron transfer interface in the cyt f-Pc complex obtained from
molecular modelling of the cyt f-Pc interaction [6]. In short,
electron tranfer between the Fe-atom in the cyt f heme and the tyrosine
83 (Y83) in plastocyanin starts with the oxidation of the Fe-atom and electron
transfer directly to the OH group or the pi-electron system of tyrosine
1 (Y1) in cyt f, or through an intermediate pyrrole ring in the cyt f heme
macrocycle. From there, the electron transfer route goes through
the Cu-coordination centre (see details in Fig. 2) following most
likely one or several molecular pathways. A few prossible routes
are discussed in greater detail in Figs. 2-4.
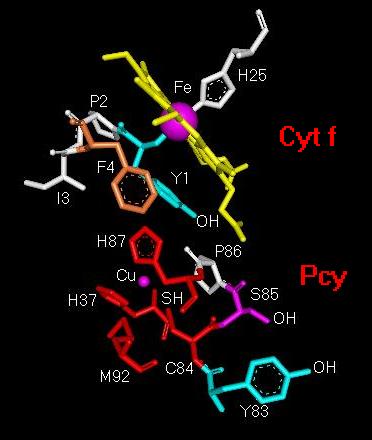
Fig. 1. Structural
detail of the molecular electron-transfer pathway from
the Fe-atom in cytochrome f (Cyt
f) to
tyrosine 83 in plastocyanin (Pcy).
Abbreviations: Cu, copper atom in Pcy; C84,
cysteine 84; Fe, iron atom in cyt f
heme; F4, phenylalanine 4; H25, histidine 25; H37,
histidine 37; H87, histidine
87; I3, isoleucine 3; M92, methionine 92; P2, proline
2; P86, proline 86; S85,
serine 85; Y1, tyrosine 1; Y83, tyrosine 83.
A schematic representation of the
distances between the amino acid residues that most likely participate
in electron transfer in the cyt f-Pc complex is given in Fig. 2.
The figure shows that the distance from the Fe-coordination
centre to Y1 is approximately 7.2 Å and 5.9 Å from Y1 to the
Cu-ion. From C84(SH) to S85(OH) and from S85(OH) to Y83(OH) the electron
transfer distances are respectively about 7.0 Å and 5.2 Å.
The figure indicates also that another possible electron transfer route
is between C84(SH) and the pi-electron system in Y83 with an interaction
distance of the order of 7.3 Å. This presents a considerable
interest since it has been quite often emphasized in past discussions (see,
e.g., [1,14]) that electron transfer distances higher than about
5 Å are a reliable indication of electron transfer occurring by tunnelling
efffect, that is, by a quantum jump from one electronic state to another.
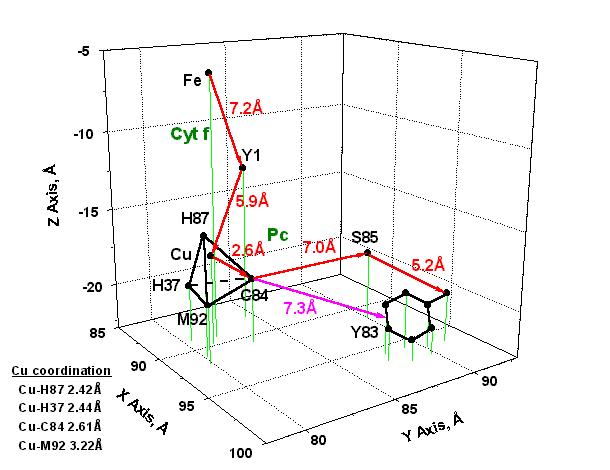
Fig. 2. Model
of electron transfer from the Fe-coordination centre in cytochrome f (Cyt
f) to the
Cu-coordination centre and tyrosine 83 in plastocyanin (Pc).
The
X, Y, Z values are those obtained
from the complex shown in Fig. 1. The most probable pathways
of electron transfer are shown with red
arrows; another possible route of electron transfer between C84
and the pi-electron system in Y83 is
indicated with a magenta arrow.
Abbreviations:
Cu, copper ion in Pc; C84, cysteine 84; Fe, iron
ion in cyt f heme; H37, histidine 37; H87, histidine 87;
M92, methionine 92; S85, serine 85;
Y1, tyrosine 1; Y83, tyrosine 83.
Fig. 2 indicates also
that the distance Cu-C84(SH) is approximately 2.6 Å, i.e., the coordination
distance from the Cu-ion to the SH group in C84. This is a quite
small electron transfer distance in relation to the 5 to 7 Å determined
for the other pathways illustrated in Fig. 2. At first, this
is a convincing argument to suggest the co-existence of different molecular
mechanisms of electron transfer in the Cyt f-Pc complex. Secondly,
an electron transfer distance of about 2.6 Å is a good indication
that in the Cu-coordination centre, and maybe also in its molecular nearness,
the electron transfer reaction originates in Van der Waals (VDW) contacts.
This question is discussed further in section (ii) below.
(ii) Structure-function model II
Fig. 2 indicates that
the Cu-ion in Pc is coordinated to the cysteine 84 (C84) thiol group, the
methionine 92 (M92) thioether group, and the histidines 37 (H37) and 87
(H87) imidazole groups in a quite highly distorted tetrahedral configuration.
The corresponding coordination distances are given in the figure.
Such unstable three-dimensional configuration has partly its origin in
differences of conformational strain energy (DHs)
of the amino acid residues (see discussion in [15]) in the Cu-coordination
centre. That is, DHs = 0.08 (C84),
11.68 (M92) and 14.15 (H37, H87) kJ/mol. This materializes in the
almost identical coordination distances for Cu-H87 (2.42 Å), Cu-H37
(2.44 Å) and Cu-C84 (2.61 Å) as compared to the quite different
Cu-M92 distance (3.22 Å) which gives thus rise to the distortion
observed in the coordination tetrahedron (see Fig. 2).
The distorted tetrahedral configuration of the Cu-coordination
centre is obviously unstable from a thermodynamic viewpoint, inusmuch as
the copper ion may adopt two different configurations; that is, the
configurations corresponding to the coordination numbers (CN) four or six
(see, e.g., [16]). It is worth noting at this point that a
fifth and a sixth coordination partners corresponding to CN = 6 are predicted,
in addition to the more common four coordination partners of the Cu-ion
in Pc which are usually identified with the amino acid residues described
above. The implicit fifth and sixth coordination partners were not,
however, correctly identified. Nevertheless, this matter raises an
interesting point. In fact, the crystal radii for the Cu-ion being
0.74 Å for CN = 4 and 0.91 Å for CN = 6 [16], it becomes
perceptible that any molecular volume change in the Cu-centre should
have functional consequences that have not yet been determined, but
which are certainly instrumental in influencing the electron transfer rate
in cyt f-Pc, at least in the route from the Cu-ion to Y83. This may
take place, for example, by changing the arrangement and the reorganizational
energy [1] of the Van der Waals contacts in the region comprising
the H87 imidazole ring, the Cu-ion and the C84 thiol group (Fig 3).
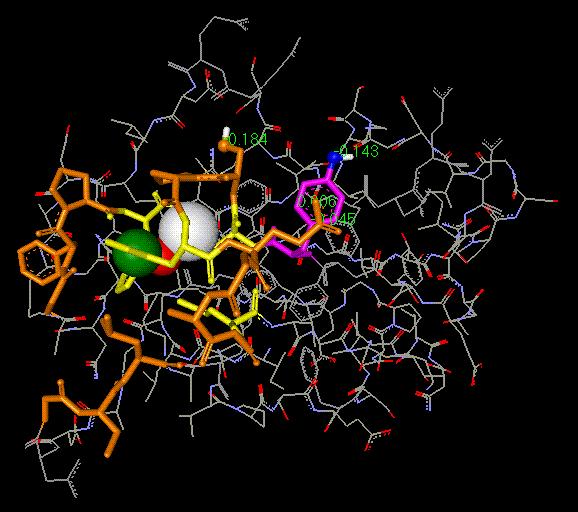
Fig. 3. Illustration
of the Van der Waals (VDW) contacts near the Cu-coordination
centre in plastocyanin (Pc). The VDW contacts are seen
in histidine 87 (olive), the Cu-
atom (red) and the S-atom in cysteine
84 (C84; white). The electron transfer beyond C84
and up to the pi-electron system of tyrosine 83 (Y83; magenta)
or the the OH group in Y83
(dark blue) takes place most likely
by tunneling effect. The amino acid residues given in
yellow colour are those participating
in copper coordination. The 'hydrophobic crown'
(dark brown) surrounding the Cu-coordination
center is also shown. Note that this
hydrophobic patch is active in the docking of plastocyanin with cytochrome
f.
From the above considerations, it is reasonable to foresee
that the transfer of an electron from Y1 (in cyt f) to the Cu-ion in plastocyanin,
the bond lengths in the Cu-coordination centre must distort from their
stable, or steady-state geometry, to another distorted geometry,
or transition-state geometry, where lenghtening and contraction
of bonds occur. For this to take place, energy is expended prior
to the return of the coordinnation centre to its equilibrium geometry.
This model is in accord with the Franck-Condon principle since the transfer
of an electron is faster than the nuclear motions, thereby meaning that
the nuclear positions are so to say 'frozen' in time during the electron
transfer process. A model to take into account these effects is presented
in Fig. 4 where the 'Van der Waals route', that is, the Y1(cyt
f)-->H87(Pc)-->Cu(Pc)
molecular pathway, is shown in parallel with the 'tunnelling effect
pathways' discussed above, i.e., (i) Fe(cyt f)-->Y1(cyt
f), (ii) Cu(Pc)[or C84(Pc)]-->S85(Pc)-->Y83(Pc),
and (iii) Cu(Pc)[or C84(Pc)]-->Y83[pi-electron
system](Pc).
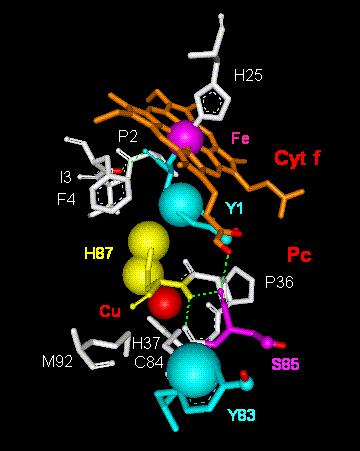
Fig. 4. Model
of electron transfer via Van der Waals (VDW) contacts throughout
tyrosine 1 in cytochrome f (Cyt
f) to histidine 87 and the Cu-atom in plastocyanin
(Pc). Abbreviations:
Cu,
copper atom in Pc; C84, cysteine 84; Fe, iron atom in cyt f
heme; F4, phenylalanine 4; H25, histidine 25; H37,
histidine 37; H87, histidine 87;
I3, isoleucine 3; M92, methionine 92; P2, proline 2;
P36, proline 36; S85, serine 85;
Y1, tyrosine 1; Y83, tyrosine 83.
4. Concluding
remarks
back
First,
the data shown in Figs. 2 and 4 indicate that different electron
transfer mechanisms may take place in the same cyt f-Pc complex, that is,
electron transfer by tunnelling effect and via Van der Waals contacts.
This may have far reaching consequences since, in a first approximation,
it presupposes the parallel function of adiabatic and non-adiabatic electron
transfer mechanisms, thereby implying the interplay of mechanisms with
different temperature dependence, or requirements [1,14].
The coexistence of such processes may well constitute a means of control
of the electron transfer activity not only in the cyt-Pc complex, but also
between the cytochrome b6f complex and P700, the reaction center of photosystem
I.
Secondly, it is noted that two other questions
still remain. The first is the possibility that water molecules may
provide an efficient pathway of electron transfer (see discussions in [17,18]).
In which the cyt f-Pc complex is concerned, the mediation or facilitation
of electron transfer by interposed water molecules inside the protein complex
is an assumption that, though attractive, has most probably to be ruled
out on account of arguments discussed in computer simulations of water-plastocyanin
interaction energies [19]. Another interesting matter is the
effect of side chain movements on electron transfer through the 'electron
transfer hole or tunnel' in the cyt f-Pc complex, which the molecular
pathways delineated in Figs. 2 and 4 tacitly predict.
A
detailed examination of these questions shall be reported elsewhere.
ACKNOWLEDGEMENTS. This work
was supported by grant OGP0006357 from the Natural Science and Engineering
Research Council of Canada and institutional grants from the Université
du Québec à Trois-Rivières, and is the result of a
collaboration with Dr. A. Kajava at the Institut Suisse de Recherches Expérimentales
sur le Cancer (ISREC), Groupe de Bioinformatique, Epalinges s/Lausanne,
Suisse. I wish to thank Dr. A. Kajava and the Bioinformatics Group
and Library staff at ISREC for the friendliness of their welcome and help,
and Dr. I. Gabashvili for many interesting comments and suggestions.
back
REFERENCES
back
[ 1] DeVault, D. (1984) Quantum-mechanical Tunnelling in
Biological Systems, 2nd ed.,
Cambridge University
Press, Cambridge.
[ 2] Friesner, R. A. (1994) Structure 2:339-343.
[ 3] Frazão, C., C. M. Soares, M. A. Carrondo, E.
Pohl, Z. Dauter, K. S. Wilson, M. Hervás, J. A.
Navarro, M. A. De
la Rosa, and G. M. Sheldrick (1995) Structure 3: 1159-1169.
[ 4] Gross, E. L. (1996) In Oxygenic Photosynthesis:
The Light Reactions. D. Ort and C. Yokum, editors.
Kluwer Academiv Publishers,
Dordrecht, The Netherlands.
[ 5] Berman, H.M., J. Westbrook, Z. Feng, G. Gilliland,
T. N. Bhat, H. Weissig, I. N. Shindyalov,
P. E. Bourne (2000)
The Protein Data Bank. Nucleic Acids Res. 28: 235-242.
[ 6] Kajava, A., and M. Fragata (in preparation).
[ 7] Roussel, A., and C. Cambillan (1989) In Silicon Graphics
Geometry Partner Directory (Fall 1989).
Silicon Graphics,
editor. Silicon Graphics, Mountain View, CA. pp 77-78.
[ 8] Roussel, A., and A. G. Inisan (1993) TURBO-FRODO,
Version 4.3, release a. Bio-Graphics,
Marseille, France.
[ 9] Kajava, A. V. (1996) Proteins: Structure, Function
and Genetics 24:218-226.
[10] Bohner, H., H. Böhme, and P. Böger (1980)
Biochim.
Biophys. Acta 592:103-112.
[11] Sandmann, G., H. Reck., E. Kessler, and P. Böger (1983)
Arch.
Microbiol. 134:23-27.
[12] Ho, K. K., and D. W. Krogmann (1984) Biochim. Biophys.
Acta 766:310-316.
[13] Zhang, H., H. B. Pakrasi, and J. Whitmarsh (1994) J.
Biol. Chem. 269: 5036-5042.
[14] Marcus, R. A., and Sutin, N. (1985) Biochim. Biophys.
Acta 811: 265-322.
[15] Sak, K., M. Karelson, and J. Jarv (1999) Bioorg. Chem.27:
434-442.
[16] Douglas, B., D. H. McDaniel, and J. J. Alexander (1983)
Concepts and Models of Inorganic
Chemistry, 2nd ed.,
John Wiley, New York.
[17] Prigge, S. T., A. S. Kolhekar, B. A. Eipper, R. E. Mains,
L. M. Amzel (1999) Nature Struct. Biol.
6: 976-983.
[18] Tripathi, G. N. R. (1998) J. Am. Chem. Soc. 120:
4161-4166.
[19] Wang, C.X., and S. Cannistraro (1985) Il Nuovo Cimento
5:
405-414.
THE END
 fragata@uqtr.uquebec.ca
fragata@uqtr.uquebec.ca
Trois-Rivières, July 5, 2000
back



